News > Pathoplexus July Update
Pathoplexus July Update
By the Pathoplexus Team - 14 July 2025
For many at Pathoplexus, summer is now in full swing (and winter well underway for many others) - and we’re happy to bring another news update! The past few months have been particularly exciting as we’ve seen an incredible increase in direct submissions from around the world, as well as the addition of some great new features - all against the backdrop of our continuous effort to improve Pathoplexus for the community.
Grab your sunglasses (or woolly hat) and we’ll dive into the latest milestones, improvements, and events!
Direct full-genome submissions:
May and June have brought with them a surge in direct uploads to Pathoplexus, across 5 of our supported pathogens, 5 continents, and 8 countries! In fact, so far in 2025, Pathoplexus has received more direct mpox submissions than INSDC.
-
We’ve received 128 mpox sequences from the National Microbiology Laboratory, Public Health Agency of Canada; Instituto Nacional de Saude Doutor Ricardo Jorge (INSA), Portugal; National Virus Reference Laboratory, University College Dublin, Ireland; National Public Health Agency (NPHA), Ministry of Health, and Kenema Government Hospital, Sierra Leone; Central Public Health Reference Laboratory and National Public Health Agency (NPHA), Sierra Leone and Africa Centers for Disease Control and Prevention; Cambodia Mpox Outbreak Surveillance and Research Team, Virology Unit, Institut Pasteur du Cambodge, Phnomn Penn, Cambodia; and the Centre de recherche de formation en infectiologie de Guinee (CERFIG), Guinea
-
For West Nile, we’ve received 15 sequences from Unité des Virus Émergents (Aix-Marseille Université - IRD190 – Inserm1207 - EFS – IRBA), France and Pathogenesis and Control of Chronic and Emerging Infections (PCCEI), Altopictus, Unite des Virus emergents (Aix-Marseille Universite - IRD190, Inserm1207 - EFS, IRBA), France
-
For RSV-A, 1,089 sequences were shared from Clinical Virology, Basel, Switzerland; National Virus Reference Laboratory, University College Dublin, Dublin, Ireland; Laboratorio de Salud Publica, Facultad de Ciencias Exactas, Universidad Nacional de La Plata, Argentina; and Institut de Recerca, Hospital Universitari Vall d’Hebron, Spain
-
For RSV-B, 1,060 sequences were shared from Clinical Virology, University Hospital Basel, Basel, Switzerland; National Virus Reference Laboratory, University College Dublin, Dublin, Ireland; Laboratorio de Salud Publica, Facultad de Ciencias Exactas, Universidad Nacional de La Plata, Argentina; and Institut de Recerca, Hospital Universitari Vall d’Hebron, Spain
-
And finally, we received 1 laboratory (not clinical) Ebola Zaire sequence from Special Pathogens, National Microbiology Laboratory, Public Health Agency of Canada, Canada
Updates on funding, collaboration, and sequences accepted:
If you’ve ever thought about donating to Pathoplexus, we’ve now added a new Funding Page to help locate useful information - and to give directly! We’ll be adding additional donation methods soon. If your organization or department would like to offer institutional support, please do get in touch!
We’re also thrilled that ARTIC2 - the next chapter of the hugely successful ARTIC project - has officially kicked-off! Pathoplexus co-founder Emma Hodcroft is part of the ARTIC2 consortium, and will focus on developing Loculus (the software underlying Pathoplexus) to integrate with ARTIC sequencing and analysis software - and make sending data from local Loculus instances to Pathoplexus easy and fast.
Finally, to reflect that Pathoplexus is focused on sharing sequences from epidemiological and biological settings for research and surveillance, our Terms of Service have been updated to highlight that we do not accept in silico or laboratory construct sequences (e.g. for patent purposes). Such synthetic sequences will be revoked. Note that we do accept vaccine-derived sequences (e.g. sequences derived from real infections that originated from a vaccination).
Technical updates:
-
Finding sequences from the lineage or clade you want is now even easier thanks to the ability to include sublineages in your search! For example in mpox, searching lineage
A.2will return 83 sequences with exactly that lineage assignment, whereas turning on “include sublineages” will includeA.2.1,A.2.2, andA.2.3in addition toA.2and return 356 sequences! In the API, you can append*- for example,A.2*orA.2.*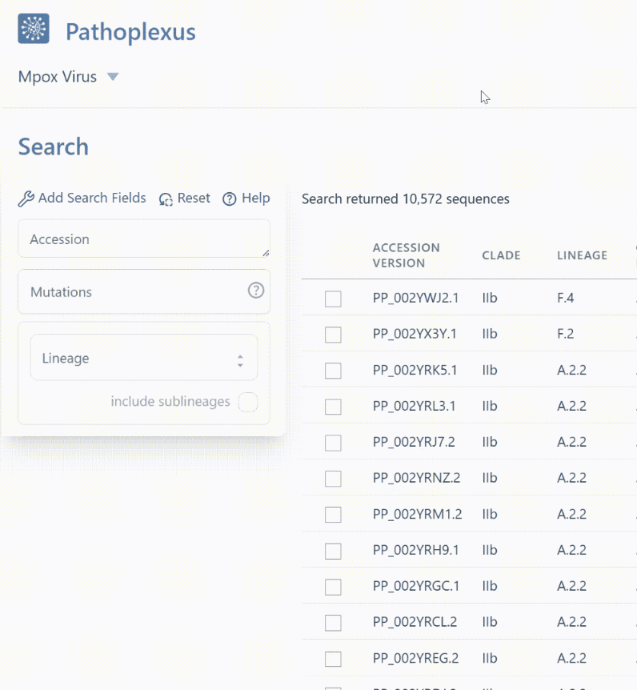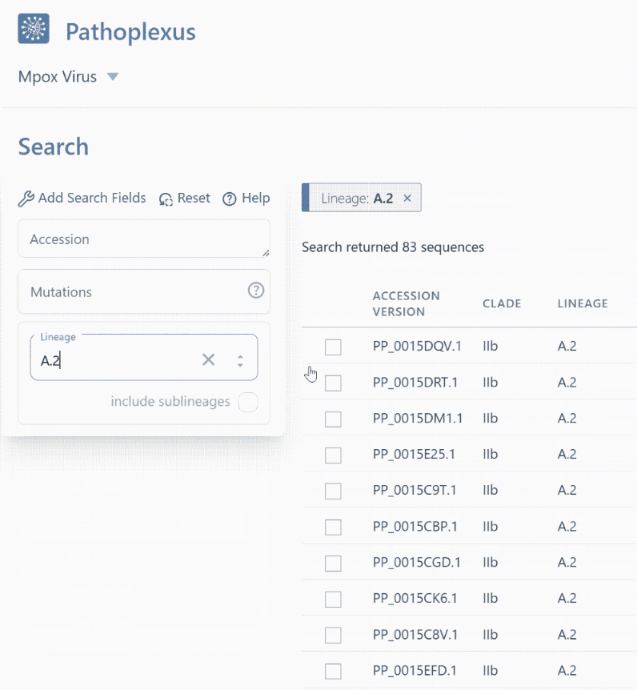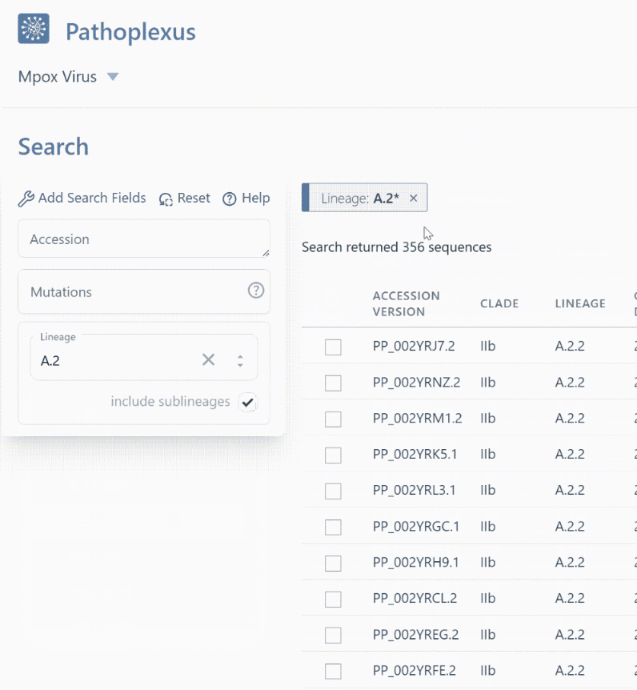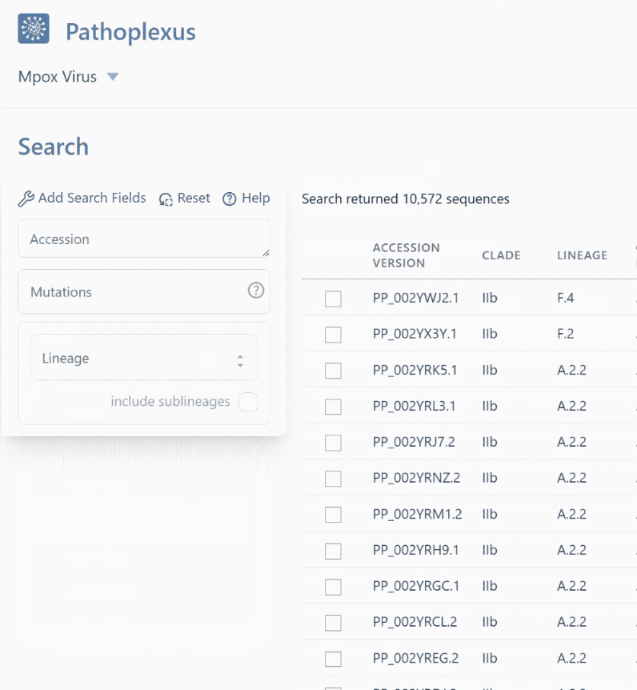
-
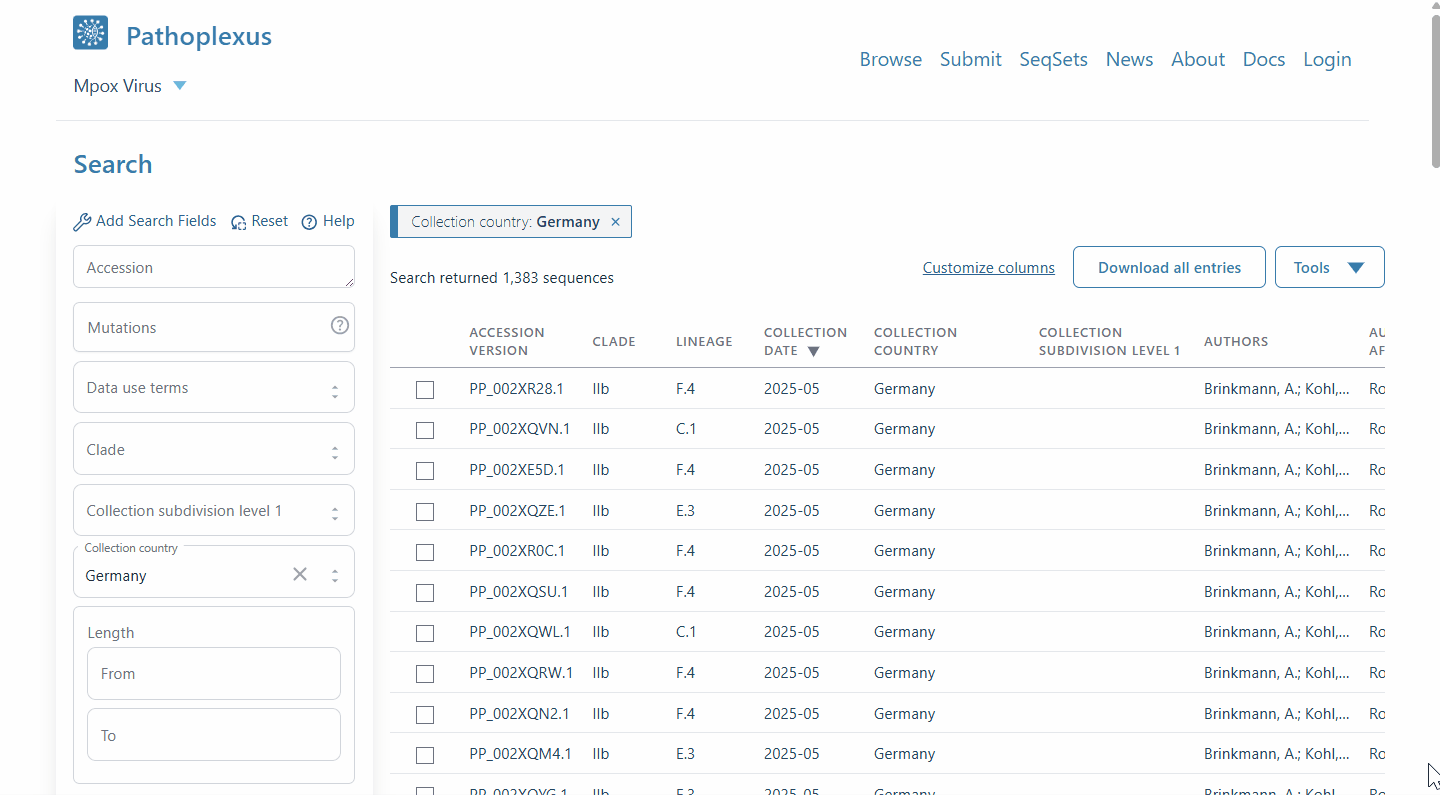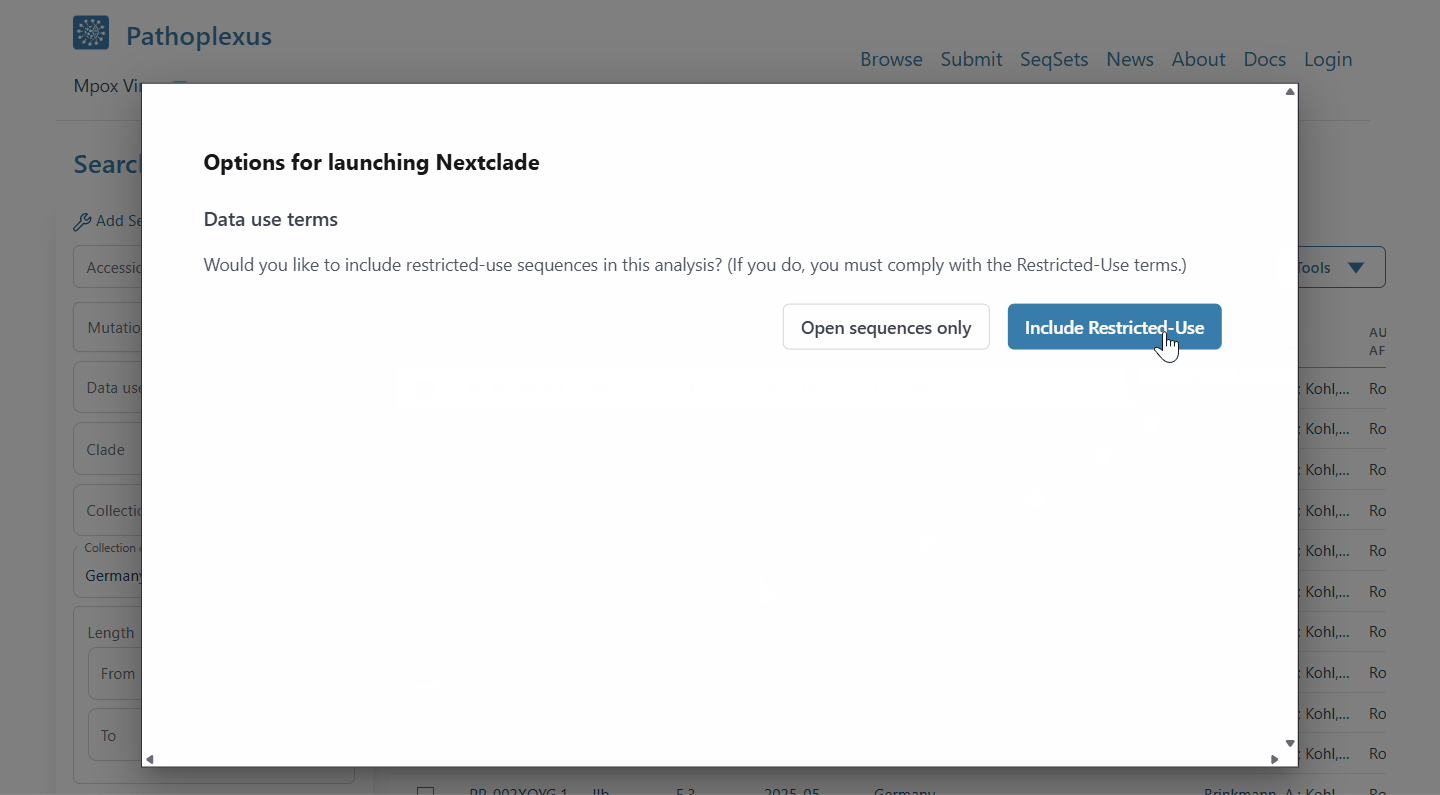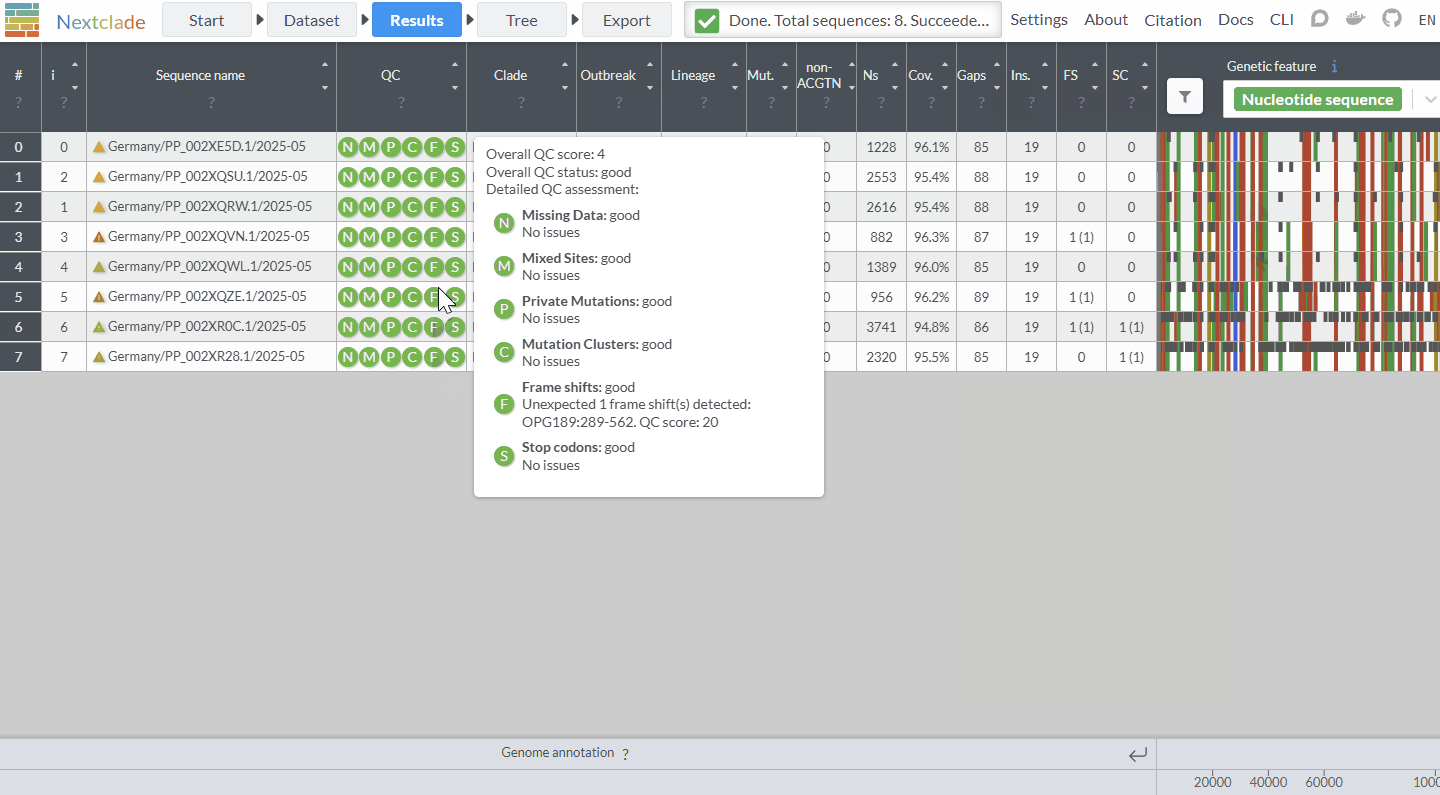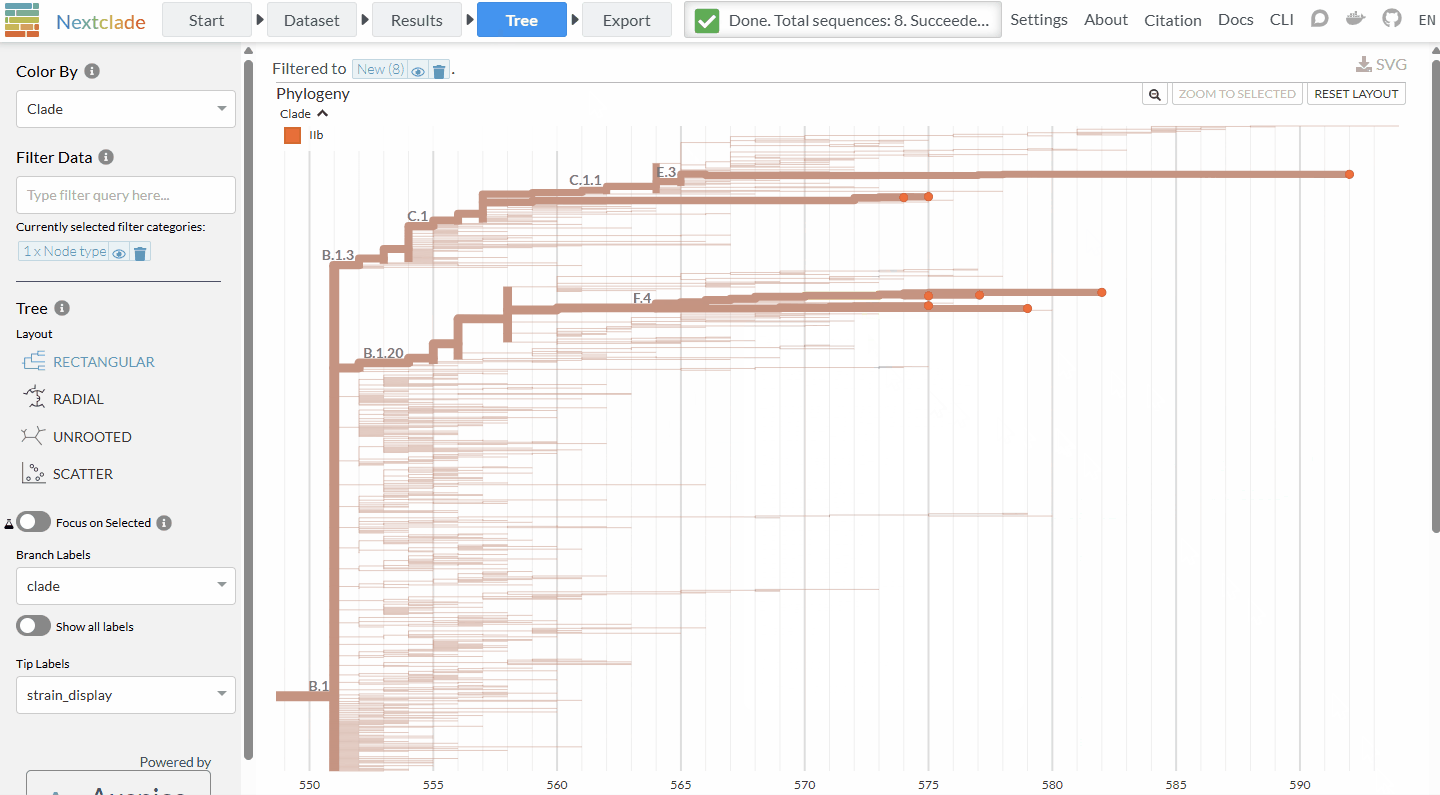
Ever wished you could quickly look at a few sequences in Pathoplexus in context - or see the distribution on a map? With our new ‘linkout’ feature (‘Tools’ on a search page), now you can! After selecting some sequences, select ‘Nextclade’ to run a quick analysis and place the sequences on a reference tree to see how they relate to each other, or ‘Country Map’ to see the distribution of sampling locations around the world. If you’d be interested in having your tool included, please see our docs!
-
When we download segmented virus sequences from NCBI Virus, it can sometimes be a challenge to link segments together as being from one sample. A recent update to NCBI Virus now also passes along a new metadata field (
strain), which has allowed us to more accurately group over 45 CCHF segments submitted from the Chinese Academy of Sciences, Wuhan and the Institut Pasteur, Dakar. -
It’s been great to see an uptick in people using our SeqSet feature! To help show off more of the information about the sequences in the SeqSet, we’ve added metadata tables to SeqSets pages. We hope to add more features to these pages soon!
-
Using the API to filter the data you need is even easier now, thanks to an update that adds support for advanced qualifiers to queries! For example:
- Logical expressions: You can now use
and,orandnot(also&,|, and!) - Support for regex: ex:
host.regex=.*bos.* - “N-of” and “exactly-of” to match sequences with at least N (or exactly N) of the desired parameters, e.g.
[3-of: mutation1, mutation2, somethingelse…]or[exactly-3-of:...] - See the full documentation on using these advanced queries here!
- Logical expressions: You can now use
In the media:
We were thrilled to see Pathoplexus co-founder and Chair of the Executive Board Emma Hodcroft featured in the University of Basel ‘UniNova’ magazine, in an interview that featured Pathoplexus, underscored the importance of working together in science, and highlighted how shifts in global science policy could impede our ability to counter infectious diseases.
Past talks:
- Emma Hodcroft spoke about Pathoplexus at at Dynamics & Evolution of Human Viruses (6-9 May, Asnières-sur-Oise, France)
- Chaoran Chen spoke about Loculus at Applied Bioinformatics & Public Health Microbiology (ABPHM) (21-23 May, Hinxton, UK)
- Emma Hodcroft spoke about data sharing with Pathoplexus at the Swiss Pathogen Surveillance Platform (SPSP) annual meeting (10 June, Bern, Switzerland)
- Pathoplexus was excited to be invited to speak about the importance of enabling fast, federated pathogen data sharing at d/acc Berlin, where Emma Hodcroft spoke (11 June, Berlin, Germany - livestream recording)
- Finally the Swiss Sarcoma Society was excited to hear about novel forms of data sharing during a talk on Pathoplexus by Emma Hodcroft (26 June, Lucerne, Switzerland)
Meetings, Minutes, and Resolutions:
- Remember that you can always check our minutes from General Assemblies and Executive Board meetings, as well as Executive Board resolutions